Maize genomics
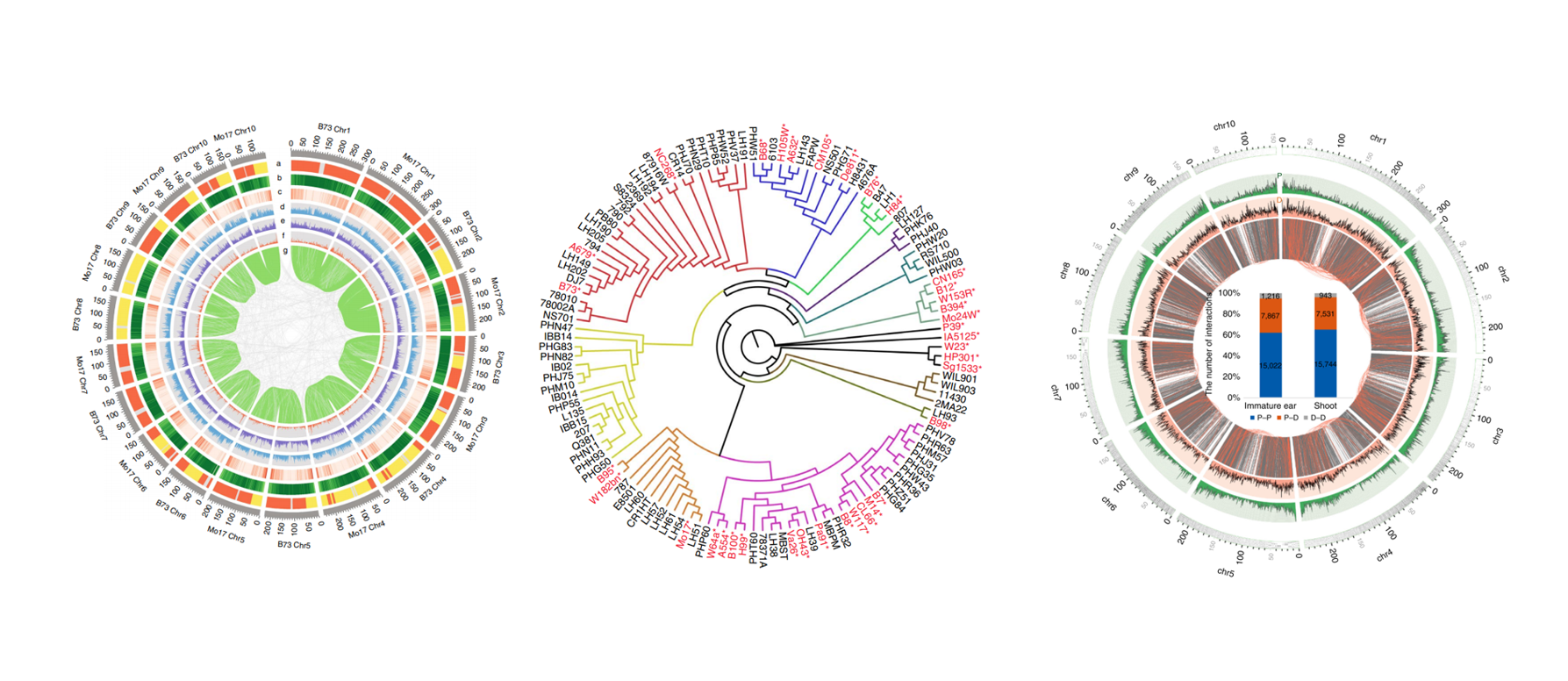
Maize is an important crop with a high level of genome diversity and heterosis. Our maize genomics studies cover maize population genetics, de novo maize germplasm assembly, and multi-omics data integration. The lab has reported a de novo genome assembly of a corresponding male representative line, Mo17 and has demonstrated the genetic changes correlated with remarkable increases in productivity during modern maize breeding. Moreover, the lab also uncovered the chromatin interaction maps in maize and investigated the relationship between three-dimensional chromatin interactions with transcriptional regulation of genes.

Maize genetic engineering and molecular breeding
We have devoted to creating new insect-resistant genetically modified maize through integrating the haplotype breeding technology, the whole genome selection technology, and the hybrid breeding technology. Our group have quickly introduced insect-resistant 2A-7 into the elite inbred lines, and bred prospective new genetically modified varieties. Our long-term mission is to build an efficient and high-throughput transgenic backcross breeding platform.
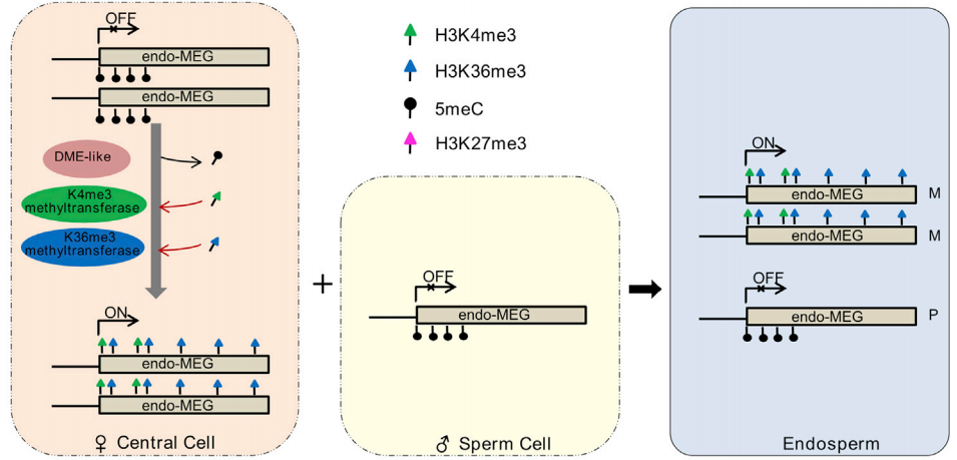
Gene imprinting in maize endosperm
Genomic imprinting refers to allele-specific expression of genes depending on their parental origin. We have uncovered the complex patterns of mutually exclusive epigenetic modifications deposited at different alleles of imprinted genes that are required for genomic imprinting in maize endosperm. In addition, we have provided the first genome-wide map of allele-specific nucleosome occupancy in plants and suggests a mechanistic connection between chromatin organization and genomic imprinting.

Maize quantitative genetics and improvement
We are focusing on isolating QTLs underlying yield and quality related traits in maize by using multiple strategies such as genome-wide association study, linkage mapping and so on. Using quantitative and population genetics approaches to understand the genetic architecture of agronomically important traits, and how different alleles shape the patterns of plant development and adaptation. Through our study, we want to improve targeted yield and quality related traits genetically via pyramiding different elite alleles.
